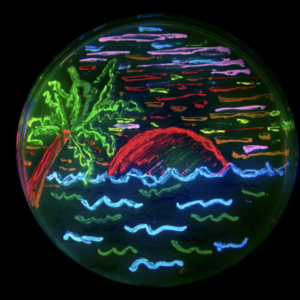
What if I told you that this colorful plate depicting a tropical landscape with ocean waves and a setting sun is not a painting, but actually consists of many bacterial colonies. Each streak on this petri dish contains a colony of bacteria expressing fluorescent proteins. These proteins have been modified to absorb and subsequently emit light at specific wavelengths we perceive as various colors. Besides making pretty petri-dish art, the discovery of the original green fluorescent protein and subsequent experimentation of the complex allowed access to a powerful and diverse tool that has revolutionized the way researchers view and study cellular processes.
History of the GFP:
In 1962, Japanese researcher Osamu Shimomura and his team accidentally discovered a green fluorescent protein while attempting to isolate the proteins that contribute to bioluminescence. Bioluminescence is a metabolic process in which an organism emits light from their body and it appears in species like fireflies and crystal jellyfish. Aequorea Victoria, more commonly known as crystal jellyfish, regularly exhibits blue luminescence, and their ease of access piqued the interest of scientists to study this phenomenon.
Bioluminescence vs Fluorescence (with info about visible light)
Fluorescence and bioluminescence both utilize various biological processes to produce light that exhibits a wide range of recognizable colors that form our rainbow, but they differ in terms of the mechanism that causes that emission of light. In bioluminescence, the process is typically a chemical reaction, while a different phenomenon causes fluorescence light. Although light exists at many different wavelengths and frequencies, we can only see light ranging from 400-700 nanometers in length, which includes the myriad of colors that we call the rainbow. Each color is distinguishable by subranges of wavelengths, for example, the color green can be observed from waves of light ranging from 495 to 570 nanometers in length. Fluorescent proteins function by absorbing light at a specific wavelength, or color, that excites the protein to subsequently emit light at a different wavelength. As such, shining blue light at a green fluorescent protein (GFP) causes it to become excited and glow green.
Diversification of the Green Fluorescent Protein
The discovery of GFPs by Dr. Shimomura prompted other biologists to tinker with its fluorescent properties. By altering the DNA sequence of the protein via genetic tools and techniques, researchers were able to tinker with protein structure that directly affects the range at which proteins absorb and emit light. This enabled the creation of other fluorescent proteins, such as the commonly used bright-red mCherry with greater resistance to changes in pH and thus a better candidate for more strenuous experiments than its green cousin, the GFP. Labs, such as the lab of Nobel Prize winning Dr. Roger Tsien, studied which mutations of the fluorescent protein yielded the most consistent and beneficial properties, including protein stability, expression, and of course, color. With advancements in genetic engineering, continuous modulation of these proteins over the past two decades has been an integral part in the creation of the field of cellular imaging.
Plasmids, Transfection and Cellular Imaging
Although DNA sequences that code for fluorescent proteins can be modified to produce different colors and characteristics, the process in which these proteins are expressed is quite unique. The DNA sequences that encode for fluorescent proteins are usually engineered within larger DNA complexes called plasmids. Plasmids are unique circles of DNA that bacteria primarily use for a process called transformation. This process allows bacteria to occasionally absorb a plasmid and its DNA contents to make use of the new genetic material. This results in the synthesis of proteins whose coding sequences are found within the plasmid and allows bacteria to rapidly adapt to new environments. Researchers utilize plasmid transformation to transfect or insert plasmids of their choice into bacteria and other cell lines. Under favorable conditions, and with a bit of luck, these plasmids enter the cells, but this phenomenon is hard to confirm with the naked eye. As such, plasmids containing fluorescent protein sequences prove to be excellent at confirming successful transfection via subsequent fluorescence post-transfection.
Localization of Fluorescent Proteins
Successful insertion of fluorescent sequences laid the foundation for the future of bioimaging. In 2002, researchers discovered that the inclusion of specific sequences directly before a protein gene can direct protein compartmentalization. In essence, these little genetic tags act as guides that lead proteins to specific areas of the cell and to avoid others. With targeting sequences and fluorescent proteins working in tandem, scientists direct fluorescent proteins to certain regions, such as the nucleus, to precisely visualize these regions of interest.
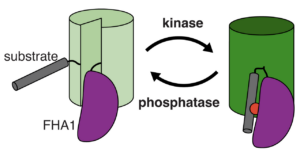
Biosensors
The discovery of subcellular compartmentalization catalyzed a revolution in the field of cell imaging and directly contributed towards the development of the first biosensors. Biosensors are multiprotein complexes that contain fluorescent proteins and subunits that detect molecules of interest. These subunits exist as a certain shape, and when they sense or bind to a specific molecule, the newly formed complex alters the structure of the biosensor as a whole. When biosensor structures are significantly altered, the change can significantly impact the fluorescence of the proteins within. Biosensors allow for dynamic visualization of metabolic pathways that were previously impossible to observe in real time without interruption of the processes themselves. As such, these multiprotein complexes’ contribution to cellular imaging have made biosensors indispensable in modern research. Scientists have continued to develop and rapidly expand biosensors in recent years, with sensors existing for nearly every metabolite you could imagine and structure varying from small complexes to large ones.
Conclusion
Over half a century has passed since the discovery of the green fluorescent protein. After worldwide recognition in Stockholm, Sweden, fluorescent protein research has come a long way from humble beginnings in the depths of the Pacific. Biosensors continue to be integral parts of modern research from utilizing multiple fluorescent proteins and sensing subunits to visualizing everything from subcellular ATP levels to calcium ion levels. With continued success in the field, who knows what biosensors will be developed and where they will lead!
Works Cited:
https://scintillon.org/Shaner.html
https://www.conncoll.edu/ccacad/zimmer/GFP-ww/shimomura.html
https://bmcresnotes.biomedcentral.com/articles/10.1186/1756-0500-3-303
https://www.sciencedirect.com/science/article/pii/B9780122007316500057
https://www.nature.com/articles/s41589-020-00718-x

