As new sequencing technologies emerge, scientists can characterize previously unknown microbes living on the shells of inside marine invertebrate hosts. UC San Diego Roy lab investigates how these microbial universes fluctuate with our ever-changing climate and what that means for their hosts and for us.
While sunbathing or surfing at La Jolla’s beaches, you may find a diverse array of marine invertebrates, ranging from barnacles to periwinkles to mussels. You might have even gone tide pooling to see what marine life you can find. However, few give thought to the invisible, yet key players in the intertidal ecosystem–symbiotic microbes.
Microbes associated with marine invertebrates, seemingly insignificant pieces of the marine ecological puzzle, still remain poorly studied due to limitations in technology. However, microbe-host interactions in vertebrate systems are increasingly well-characterized: in particular, we know that the human microbiome plays an essential role in our digestion, immunity, and stress mechanisms.¹ However, the mechanisms by which the microbiome of invertebrates, such as mussels and sea snails, affects their host is not yet clear.² Using what we know about microbe-vertebrate interactions, researchers can infer that microbes greatly influence the physiology and metabolism of marine invertebrates as well.
In particular, it is important to explore how these microbial-invertebrate interactions are affected by environmental factors such as climate change.² If environmental changes cause microbial systems to divert from their normal distributions, what would happen to their invertebrate hosts? All current models of ecosystem responses to anthropogenic, or human-caused, climate change only factor in plant and animal interactions, excluding microbial interactions. To create a more accurate model, we need to evaluate the ecosystem holistically by including how bacteria respond to climate change and other anthropogenic stressors. Otherwise, there is potential for errors in the predictions of climate change effects on invertebrate hosts. This limitation of current models could lead to many consequences beyond the biology itself–for example, communities that depend on these fisheries for their livelihood could be negatively affected.
Biofilms: Microbial Cities
The unique composition of microbial species living in a region of an organism is known as a microbiome. In invertebrate organisms with a shell, one of the crucial microbiomes consists of a shell surface biofilm. Biofilms are a conglomeration of microbes bound by a slimy extracellular matrix that enables them to bind to foreign surfaces. When a bacterium adheres to a surface, it secretes a polymeric mucus that recruits other bacteria, protozoans, and even eukaryotes to adhere to the matrix and form a biofilm.
Biofilm communities are crucial to the wellbeing of an ecosystem. Primarily, biofilms can protect microbes from environmental threats faced by free-living bacteria. Additionally, biofilms increase microbial access and absorption of nutrients. The invertebrate’s shell surface biofilm has a host-specific, unique microbial composition different from that in marine water and sand. This specificity of microbial composition is determined by various levels of host regulation, such as nitrogen excretion, oxygen metabolism, and symbiotic feedback of the microbiome feeding into its host.³ If some of these environmental conditions change, it is hypothesized that the microbes that depend on such aspects are also affected and subsequently react to the change.
Dr. Kaustuv Roy’s lab in UC San Diego’s Division of Biological Sciences has investigated the biological and physical processes that generate large scale gradients in marine biodiversity for the past 30 years. Currently, the lab is investigating the impact of anthropogenic climate change on microbiome-invertebrate host systems using genomics, environmental data, and geographical mapping. Dr. Roy’s work is focused on understanding how changes such as nutrient availability, temperature and pH affect the distribution of the microbial communities growing on these marine invertebrates.
Latitude and Diversity
One way to probe these environmental factors is to investigate how microbe communities on marine hosts differ with geographical latitude. The latitudinal diversity gradient proposes that animal and plant species diversity is generally highest in the tropical areas and declines towards higher latitudes. To investigate whether the same is true for microbiome diversity, the Roy lab chose the invertebrate host model of Mytilus californianus, or the California mussel. This mussel has a large geographic distribution from Baja California to southern Alaska and is abundant in the intertidal zone, making it easy to find and sample. Additionally, marine bivalves such as mussels follow the common trend of higher diversity near the equator with a decline towards the poles. To identify whether the microbes on the host followed the same trend, the lab gathered biofilm samples by swabbing the gills and shells of the mussel at various latitudes ranging from Alaska to La Jolla.
Generally, microbes in a large sample are identified by genomic sequencing. To do this, particular regions of the bacterial genome are compared across various species. Microbiologists commonly use the 16S ribosomal RNA (16S rRNA) gene sequence to identify bacterial species. As a component of ribosomal RNA, this gene is essential for microbial survival as it plays a crucial role in protein synthesis. For these reasons, the 16S rRNA gene is evolutionarily conserved throughout all prokaryotes. Specifically, it consists of highly conserved regions, shared across all bacteria, which are interspersed with variable regions that closely correlate with taxonomy grouping and can be used to identify particular species of microbes. Amplification and sequencing these variable regions can thus allow us to compare and identify the various microbes through sequence homology to databases of known species.
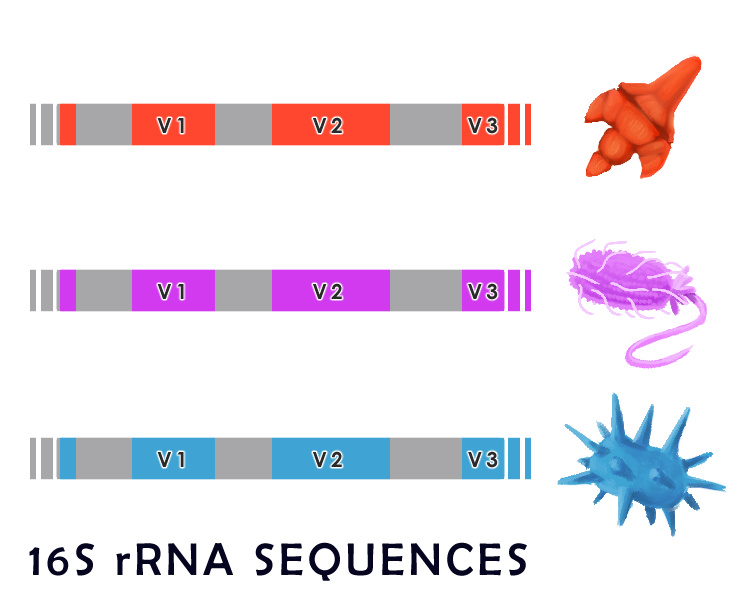
Though large-scale genetic sequencing is a powerful methodology to identify short sequences from a pooled sample, this technology has several drawbacks. Since databases of poorly studied and non-model organisms are incomplete, and only short segments are sequenced in 16S amplification, microbes can often only be identified to the phylum level.
The Roy group gathered 103 California mussel shell and gill samples at various latitudes, generating more than 13,000 variable sequences. These sequences were analyzed against a database of the 16S rRNA sequences of known species, and via sequence homology comparison, the possible types of microbes that were present on the shell at various latitudes could be narrowed down and identified.
The Roy group discovered that host-associated microbes on mollusk shell surfaces do not follow the same latitude diversity gradient displayed by marine invertebrate hosts. In fact, the microbiome on the shell tends to display an inverse latitude diversity gradient, becoming more diverse in mid or high latitudes rather than further south. An increase in the species pool of free-floating bacteria in the ocean which correlated with an increase in latitude could explain the increased taxonomic diversity of the shell surface microbes. Additionally, other environmental factors such as temperature, ocean pH, and host characteristics–all of which change with latitude–could explain the microbial diversity trends.
On the other hand, gill community diversity had no significant trend with latitude. These results suggest that environmental factors that change with latitude play a smaller role than local conditions, such as host physiology and diet, on the gill microbiome of the California mussel, while shell microbiome composition is regulated by a different set of variables.
Time Will Tell
Additionally, there is a demonstrated need to understand microbial communities’ temporal responses to environmental changes in temperature and ocean pH, so we can predict how biological systems will react to future changes in each respective factor across the globe. Currently, it is known that some hosts have evolved a tight symbiotic relationship with their microbes. As the climate continues to drastically change, this symbiosis creates possibility for co-extinction, where a host and its symbiont go extinct at the same time.² As a result, in a process known as “dark extinction,” we might lose microbial diversity when a host goes extinct before being documented or studied.
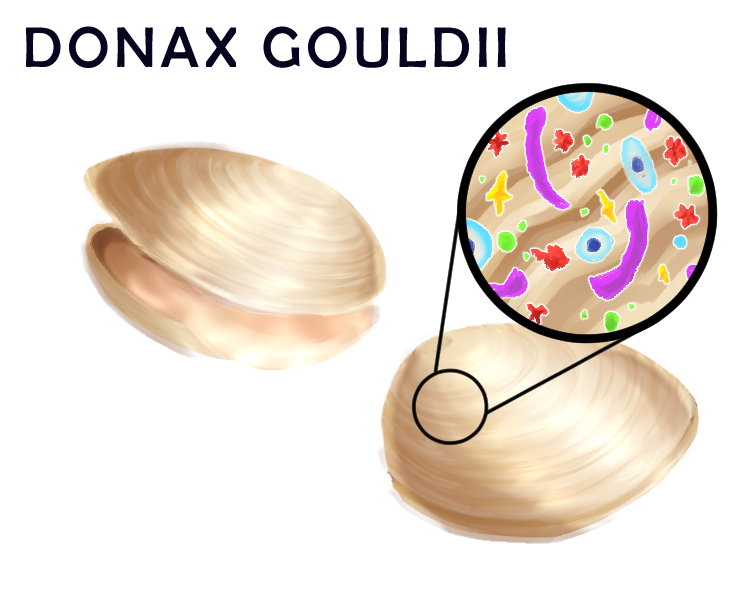
To address how microbial communities on hosts change as a result of local climate fluctuations created over large spans of time, the lab carried out a temporal study at the local La Jolla Shores beach. Researchers in Dr. Roy’s lab and at the Scripps Institute of Oceanography (SIO) analyzed the microbial diversity of samples of Donax gouldii, a small seawater clam, dating back to 2008. From the samples analyzed, it was concluded that microbial species diversity, quantified by the number of unique species present, was maintained following local environmental fluxes such as temperature changes and algal blooms. However, the identity of species constituting this constant level of biodiversity present on the hosts was completely different from year to year, with a constant turnover of microbes. Although the mechanism for how the clam keeps stable biodiversity is unclear, it is possible that the host metabolism, such as its immune system and space limitation, could be a factor.²
The Two Sides of Panama
Another experiment investigating the effect of the environment on microbe-host interactions examined microbial biodiversity in Panama, a prime location for marine ecologists because of its unique geography. In particular, the nutrient-rich Pacific Ocean and less nutrient-rich Caribbean are separated by the Isthmus of Panama, a land mass formed 3.5 million years ago between the two bodies of water. As a result, divergent evolution from the same parent species created invertebrate groups on either side of the isthmus that differed based on the oceanic environment they occupied.
Samples from seven different marine snails were collected on opposite sides of the Isthmus of Panama, and the host-microbes were sequenced and identified. Surprisingly, microbial taxa associated with hosts do not show the expected pattern of divergence following the rise of the isthmus, unlike their hosts which shared more recent common ancestors. Dr. Roy hypothesizes that because many marine host associated microbes tend to be geographically restricted, individual microbial species were not widely distributed across Panama even before the Isthmus formed. So when a land mass formed to isolate the Pacific and Atlantic oceans, the microbes associated with host populations on each side of the isthmus were already different. This study was a good example of how evolutionary divergence trends in eukaryotic hosts may not always align with those seen in their microbiomes.
More Questions than Answers
While the Roy lab has observed that various climate factors affect the microbiome, how specific factors impact microbe community composition remains to be explored. Through manipulation of temperature and pH in a tank, researchers can observe changes in invertebrate microbial composition in a controlled environment.
Additionally, the Roy group may explore the impact of the structure, texture, and material of the shell on microbial biofilm formation. How do environmental factors such as ocean acidification affect invertebrate shell surface biofilms? Working with other labs in the Jacobs School of Engineering and SIO, the group plans to investigate the effect of different shell materials and textures on biofilm formation.
This research also has implications for marine fouling, where certain microbial colonization of boat and underwater pipe surfaces recruits organisms such as barnacles to attach themselves to these surfaces. To prevent biofouling, structures are often painted with anti-biofouling paint, commonly toxic to marine life. By studying the interaction between invertebrate larvae and microbes during biofilm formation, methods to prevent biofouling without marine toxins could be elucidated.
The microbiome of an organism is a dynamic, ever-changing community, and we are just starting to explore how microbial communities associated with marine invertebrates function. Emerging technologies such as long-read sequencing platforms will soon allow scientists to sequence longer genomic segments to identify bacteria species with greater specificity, producing a finer insight into how microbial communities change over time with respect to climate changes. As researchers grow more mindful of the importance of studying large time-scale perturbations of climate, the Roy lab is archiving microbial samples in the university collection to provide resources for those who want to do a temporal study on mollusk microbiota a decade down the line. The microcosmos on the surface of marine invertebrates still has a wealth of information to divulge, and maybe it’ll be you who makes the next discovery.
References
- Carabotti, M.; Scirocco, A.; Maselli, M. A.; Severi, C. The Gut-Brain Axis: Interactions between Enteric Microbiota, Central and Enteric Nervous Systems. Ann Gastroenterol 2015, 28 (2), 203—209.
- Neu, A. T.; Hughes, I. V.; Allen, E. E.; Roy, K. Decade-scale Stability and Change in a Marine Bivalve Microbiome. Mol Ecol 2021, 30 (5), 1237—1250.
- de Carvalho, C. C. C. R. Marine Biofilms: A Successful Microbial Strategy With Economic Implications. Front. Mar. Sci. 2018, 5, 126.
- Neu, A. T.; Allen, E. E.; Roy, K. Do Host-associated Microbes Show a Contrarian Latitudinal Diversity Gradient? Insights from Mytilus Californianus , an Intertidal Foundation Host. J Biogeogr 2021, 48 (11), 2839—2852.
- Johnson, J. S.; Spakowicz, D. J.; Hong, B.Y.; Petersen, L. M.; Demkowicz, P.; Chen, L.; Leopold, S. R.; Hanson, B. M.; Agresta, H. O.; Gerstein, M.; Sodergren, E.; Weinstock, G. M. Evaluation of 16S RRNA Gene Sequencing for Species and Strain-Level Microbiome Analysis. Nat Commun 2019, 10 (1), 5029.
- Neu, A. T.; Torchin, M. E.; Allen, E. E.; Roy, K. Microbiome Divergence of Marine Gastropod Species Separated by the Isthmus of Panama; preprint; bioRxiv.
Written by Annika So
Annika is a Biochemistry Major from John Muir College. She will be graduating in 2023.
