The structure and function of proteins in the human body is largely limited by the number of amino acids that are naturally generated by the genetic code. Out of 64 codons, 61 of them code for only 20 amino acids, and the other three function as stop codons. This severely limits the proteins that our bodies can create, as well as their different functions. This is why finding ways to integrate artificial amino acids, or noncanonical amino acids (ncAAs), into the human body is so crucial, as they have the potential to enhance and expand protein structure and function. The quadruplet codon system⁸ is an innovative concept that aims to expand our original genetic system to consist of 256 codons instead of 64. This paper will discuss the development of the quadruplet codon system, alternative methods to the system, and the challenges that may come about in utilizing quadruplet codons, along with potential solutions.
Although over 500 amino acids have been found in nature,¹ only 20 are used to encode proteins within the human body. However, over the past 30 years, scientists have incorporated amino acids that do not naturally occur within the human body, known as non-canonical amino acids (ncAA), into human-made proteins. One of the earliest displays of the incorporation of ncAAs into proteins was via a suppressor tRNA with a stop codon as its corresponding codon. The tRNA codes for an ncAA, which is then incorporated into the enzyme beta-lactamase in place of the stop codon.² At the time, this field and method were relatively novel and paved the way for similar experiments.
A contemporary example of the use of ncAAs is in gene editing, a process where an organism is genetically modified to be dependent on an ncAA by introducing it into an essential gene. This also calls for an orthogonal translation system, a system containing the engineered enzyme needed to attach an ncAA to its corresponding tRNA. Modifying organisms such as viruses in this way means they cannot interact with regular cells. This doubles as an advantage for vaccination, because when using a genetically modified organism-based vaccine, horizontal gene transfer cannot occur between different systems. This means the virus can merely infect, not replicate. An immune response can then be successfully carried out.³
Another use of ncAAs includes changing the functions of certain proteins. One group took cinnamycin, a peptide antibiotic made by strains of the bacteria Streptomyces, and incorporated different ncAAs at various positions, testing out the effects of each of them. They found that with some of the new incorporations, the antibiotic activity of the now altered cinnamycin was increased⁴. Other uses of ncAAs include cancer therapy,⁵ post-translational modifications within proteins,⁶ and more.
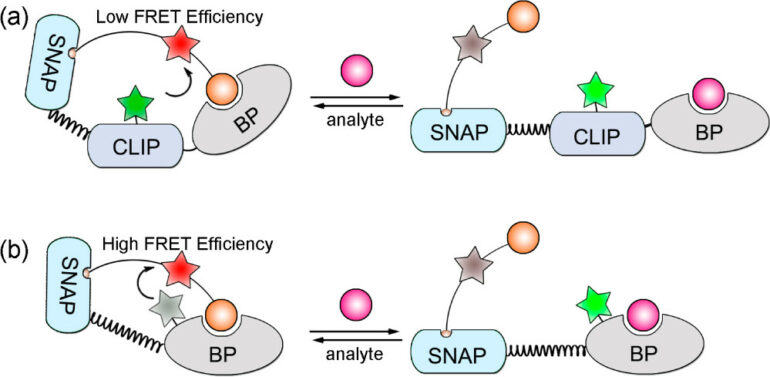
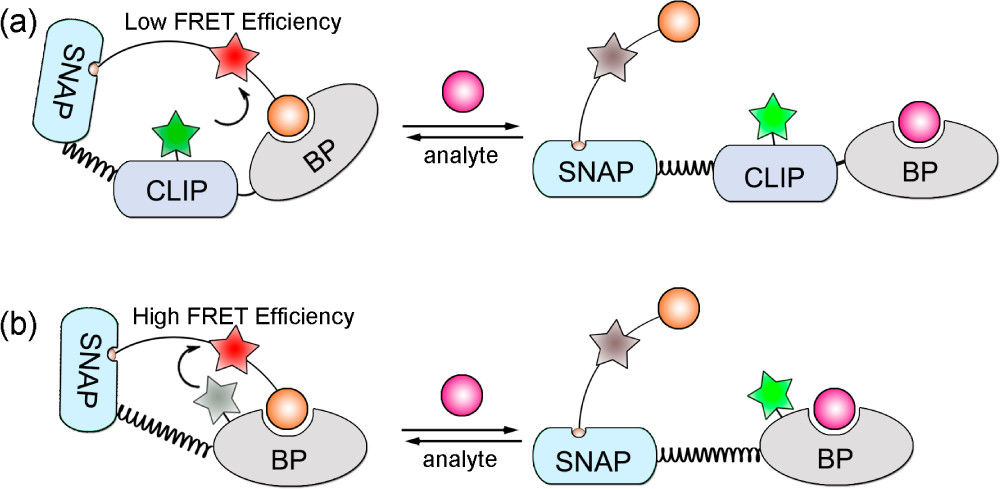
A more intricate application includes ncAA use in biosensors, specifically involving a phenomenon known as fluorescence resonance energy transfer (FRET) (Figure 1). In FRET, energy is transferred between two fluorophores—a donor molecule and an acceptor molecule, both typically fused to a binder protein. The closer these two molecules are to each other, the higher the FRET efficiency. Initially, researchers used a Snifit, which is an indicator consisting of a SNAP-tag (a self-labeling protein tag), a CLIP-tag (a fluorescent protein which can be tagged with a fluorophore), and a binding protein (BP). Afterwards, they replaced the CLIP- tag with a fluorophore-tagged ncAA, which is much smaller than the CLIP-tag. This decreased the distance between the two fluorophores, which increased FRET efficiency. These new Snifits were labeled uSnifits, and the scientists found that the dynamic range of the sensors increased. They also found that the labeling process could be carried out in vivo when using an ncAA.⁷
In order to incorporate new ncAAs into humans, however, it is necessary to develop a way to expand our genetic code. Multiple methods exist for this purpose. However, one of the most promising methods to appear in recent years is known as the quadruplet codon system. This system introduces new ncAAs into the body by creating and utilizing codons consisting of four nucleotides rather than the traditional three. This novel approach increases the number of possible codons by four times the original amount, giving the body more diversity when creating proteins. The main problem with this approach is that the body’s natural machinery is engineered to process triplet codons, not quadruplets. When a ribosome reads a strand of RNA, the ribosome will often select a tRNA which corresponds to a triplet codon instead of a quadruplet codon, incorporating a canonical amino acid instead of an ncAA. To counter this, scientists have been working on engineering orthogonal (meaning that it cannot interact with the natural machinery of the cell) ribosomes that read for quadruplet codons.²⁴ Along with the prospect of 256 codons as opposed to 64, the quadruplet codon system is shaping up to be one of the best candidates for the incorporation of ncAAs in vivo.
Quadruplet codons have been the subject of many experiments since the 1970s.⁹ One of the first influential studies in the field dates back to 1981, when a group of scientists experimented with the concept of frameshift suppression where frameshift mutations are overcome and a suppressor tRNA is able to read four nucleotides instead of three. The isolation of a novel suppressor, dubbed sufJ, revealed its ability to read exactly three different four-base codons upon recognition of the ACC triplet in mRNA: ACCA, ACCU and ACCC.¹⁰ Although frameshift suppression has been tested before,⁹, ¹¹, ¹² previous suppressors only worked for one or two codons of a single base, with minimal base changes. However, sufJ had low efficiency in decoding codons, around 1-2%.
In 2000, one group of scientists managed to engineer a tRNA in E. coli such that its anticodon loop was expanded by one extra nucleotide—specifically, the tRNA Su6, which contains the anticodon for the stop codon UAG and is a mutation of the tRNA used for leucine. Altering this tRNA allowed it to decode the codon UAGA with an efficiency of 13-26%, which was ten times more efficient than frameshift suppression.¹³
Further experiments were performed in the E. coli strain MRA8, as it has a temperature-sensitive release factor 1 (the protein needed to terminate translation in E. coli), which reduces competition between termination of translation and decoding of quadruplet codons. Variants of Su6 were created to assess the decoding efficiency of one UAGA codon, two UAGA codons in tandem, and a UAGA and a UACA codon. The former two were found to have an efficiency of 40%, meaning that they were correctly decoded 40% of the time. The latter had an efficiency of 10%. The major problem lay with competition with canonical amino acids. Since this tRNA was a mutation of the tRNA assigned to leucine, that amino acid was found to be a product of translation around 50% of the time.¹³
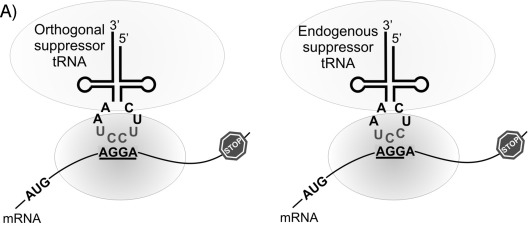
The past few decades have brought about significant findings in overcoming competition from triplet codons, eventually continuing to the engineering of orthogonal tRNAs.¹³ In 2004, J. C. Anderson et. al. created an orthogonal tRNA and aminoacyl-tRNA synthetase (aaRS) pair that was able to incorporate an ncAA, L-homoglutamine, in response to the quadruplet codon AGGA, specifically in E. coli. It was challenging to engineer an aaRS that would be specific to the ncAA, and not to any endogenous amino acids. For this reason, E. coli tRNA could not be used, because the tRNA had to be orthogonal in the organism. The tRNA was based on a prokaryotic tRNA for lysine from the bacterium Pyrococcus horikoshii. The crystal structure of its aaRS was available for study, giving a base from which the artificial aaRS could be derived. Plus, this particular bacterium is tolerant to the introduction of new nucleotides into its tRNAs.
The group performed further experiments in order to test the simultaneous incorporation of two ncAAs using a second orthogonal tRNA and aaRS pair. This pair was derived from the tRNA for tyrosine, from the organism Methanococcus jannaschii, for the artificial amino acid O-methyl-L-tyrosine. The two pairs were tested and found to be mutually orthogonal, meaning that one aaRS could not work with the tRNA from the other pair. Hence, these two were able to work together to concurrently incorporate two ncAAs into the same protein, representing a major breakthrough.¹⁴
Another group that worked on the simultaneous incorporation of two ncAAs was Hankore and colleagues.¹⁵ They decoded two quadruplet codons, UAGA and AGGA, then used them to simultaneously introduce two ncAAs into a single protein within E. coli. This group also derived their tRNA and aaRS pairs from organisms similar to those in previous studies—Methanocaldococcus jannaschii and Methanosarcina barkeri. These pairs were mutually orthogonal to both the natural amino acids within E. coli as well as each other. However, they found that the original pair derived from M. jannaschii had to be further altered through directed evolution in order to achieve greater decoding efficiency. Eventually, this was observed in certain mutant versions of the original tRNA and aaRS pair. With both the derived tRNA and aaRS pairs, they were able to successfully perform site-specific simultaneous incorporation of two ncAAs in response to two quadruplet codons.¹⁵
In 2013, researchers conducted further experiments on an orthogonal tRNA and aaRS pair, creating mutated versions of each to see which one had the highest decoding efficiency for the quadruplet codon AGGA (and thus, the highest efficiency in incorporating ncAAs), within E. coli. This experiment was significant as they found that these mutated versions also operated well in mammalian cells. This meant that the tRNA and aaRS pair used in E. coli, a prokaryote, could also be used in mammalian eukaryotic cells.¹⁶
Although many of these experiments were often tested using only one or two quadruplet codons, it is likely that these approaches or similar ones can be used for many different quadruplet codon combinations. Research has shown that site-specific incorporations within proteins allow specific, targeted changes in structure or function. The simultaneous incorporation of ncAAs also shows two mutually orthogonal tRNA and aaRS pairs working together. The studies conducted thus far demonstrate the potential of the quadruplet codon system, not only for specific incorporation of ncAAs, but also for expanding the genetic code overall through the integration of new codons into certain organisms.
While the quadruplet codon system is a promising method of expanding the genetic code, other aforementioned methods also exist for this purpose. The first method is called amber suppression. This method refers to using an orthogonal tRNA and aaRS pair in response to the amber stop codon, UAG, to insert an ncAA instead of terminating translation, which would extend the polypeptide. However, it also extends to the other two stop codons: the ochre codon (UAA) and the opal codon (UGA). In 2010, a group of scientists demonstrated in vitro incorporation of ncAAs using the three stop codons, as well as engineering tRNAs from ones existing in E. coli and yeast. Modifying the tRNA used for cysteine allowed them to insert a modified version of cysteine in response to these stop codons.¹⁷
Another group in 2010 converted the UAG codon from a stop codon to a sense codon in E. coli by removing its release factor to eliminate the possibility of translation termination. However, to avoid any harmful effects of codon alteration, the E. coli strain had to be genetically modified. It was found that the UAG codon still retained the function of stopping translation, as well as that of incorporating an ncAA, before the removal of the release factor. This problem falls under the “ambiguous intermediate” theory, where despite reassigning the genomic function of a codon, the codon still retains its previous function while shifting to the new one, prior to the elimination of the release factor. To prevent this from happening, they replaced a certain amount of UAG codons with UAA codons, to make sure that the UAG codon only had the function of incorporating an amino acid.¹⁸ A similar result was found by O’Donoghue and colleagues¹⁹ who used E. coli with the orthogonal tRNA and aaRS pair for the ncAA pyrrolysine, which works in response to UAG. They found that even though it is the only known pair to have naturally evolved to insert pyrrolysine in response to a stop codon, it cannot completely overcome the competition between this reaction and the normal termination response.¹⁹
This method of stop codon suppression is advantageous because existing codons can additionally be used for their natural functions. However, amber, ochre, and opal suppression gives us a mere three codons to work with, whereas the quadruplet codon system provides the promise of many more opportunities for ncAA incorporation because of the variety of codons that can be created. Also, when taking into consideration the fact that one codon is still required to stop translation, only two stop codons are left to assign to ncAAs. Another disadvantage arises from competition among the release factors that terminate translation. In order to experiment with amber suppression, it is necessary to mutate the bacterial strains or alter the organisms such that their release factor is not present. However, it has been found that removal of release factors can lead to a decrease in the fitness of the cell,²⁰ as ribosomes stall upon encountering the stop codons intended to be recognized by the release factors. Additionally, reassigning the function of amber codons by replacing some or all of them with UAA codons can lead to off-target mutations.⁸
The next method for expanding the genetic code comes from reassigning sense codons. In humans, 64 codons make up the genetic code, but three to four codons are generally assigned to the same amino acid. As a result, these codons only correspond to 20 amino acids. This is probably to account for possible mutations in our DNA. The codons assigned to one amino acid are very similar, so the chances that a nucleotide change will alter the amino acid are low. Reassigning sense codons involves employing infrequently utilized sense codons and having them code for an ncAA. For example, a group in 2014 reassigned the AGG codon in E. coli to code for ncAAs using the pyrrolysine tRNA and aaRS pair. This codon normally codes for arginine, so arginine would sometimes be inserted in response to AGG instead of an ncAA. The group was unable to resolve this issue.²¹ Reassigning sense codons presents many challenges and disadvantages, such as the aforementioned “ambiguous intermediate” theory. Another is an increase in the likelihood of mutations, because in the case of only one codon per amino acid, even a single nucleotide change would cause a change in amino acid.
A lesser-known method to incorporate ncAAs involves the creation of new nucleotides. A group in 2019 was able to synthesize two artificial nucleotides, dNaM and dTPT3, creating an unnatural base pair (UBP) which could then be used to create additional sense codons in a genetically modified organism (GMO), also known as a semi-synthetic organism (SSO). Out of nine new codons that were identified as stable within the DNA, three were found to be mutually orthogonal (orthogonal with respect to one another) within the SSO, increasing the total number of codons that could be decoded to 67. However, they discovered that codons with the UBP in the first position were decoded inefficiently compared to those with it in the second or third position.²² Another obstacle arises in that much of the materials required for replication, transcription, and translation have to be imported into the organism of interest via specific transporters. Each of these methods have their fair share of challenges, many of which can be overcome with the quadruplet codon system.
The quadruplet codon system has unique advantages which bypass many of the issues with the alternate solutions described, like stop codon suppression or reassigning codons. It consists of four times the number of codons that are originally available in the human body, does not need to reassign existing codons to other functions, and does not require processes that would damage the cell such as the deletion of release factors. However, a significant problem with this system is competition with triplet codons. This means that the ribosome will use tRNAs that respond to triplet codons and code for canonical amino acids, instead of using orthogonal tRNAs that read quadruplet codons and incorporate ncAAs.²³
Since the ribosome and the tRNAs that respond to triplet codons are part of the natural machinery of cells, it is difficult to incorporate quadruplet codons. This problem was first approached in 2005, when two scientists designed an orthogonal ribosome by duplicating ribosomes from E. coli. The scientists modified the ribosomes such that they would read orthogonal mRNAs (which could not be read by native ribosomes) in conjunction with the native ribosomes decoding the native mRNAs.²⁴ However, these ribosomes were not suitable when it came to incorporating ncAAs.²⁵
In 2007, Wang and colleagues expanded on the idea of orthogonal and natural ribosomes working in tandem.²⁶ An orthogonal ribosome known as ribo-X, as well as an orthogonal mRNA, and an orthogonal tRNA and aaRS pair were used, all within E. coli. Using the amber codon, they were able to increase the efficiency of the site-specific incorporation of ncAAs from 20% to 60% for a gene with one amber codon, and 1% to 20% for a gene with two. Suppressing the amber codon prevented the release factor from terminating translation without having to remove it from the cells and potentially harm the bacterium. In fact, it is hypothesized that the basis for this increase in efficiency was decreased interaction between ribo-X and the release factor. The group also speculated that it may be possible to use the orthogonal ribosomes and mRNAs to create new genetic code by using tRNAs that work well specifically with the orthogonal ribosomes.²⁶
In 2010, another group of scientists, most of whom worked on ribo-X, developed another orthogonal ribosome, ribo-Q1, which could translate an orthogonal mRNA.²⁷ This ribosome was not only able to decode the amber codon but was also able to decode several other quadruplet codons. By using an orthogonal tRNA and aaRS pair, the group was able to incorporate ncAAs in response to two of these new quadruplet codons.²⁷ These experiments summarize many of the major studies that have been done in regards to orthogonal ribosomes, which are proving to be an extremely viable solution to competition between triplet and quadruplet codons.
The quadruplet codon system has come a long way since it was first tested. Many major developments have been made to overcome potential setbacks and implement the system into mammalian cells. It can be used to introduce a multitude of ncAAs into natural systems, whose applications include, but are not limited to, gene editing, cancer therapy, vaccination, and post-translational modifications of proteins. This system is very new in the field of protein research, and much of it has yet to be explored. Still, it has high potential to expand the genetic code. Although its limitations are being challenged, scientists are researching ways to overcome them through experimentation. Many new ncAAs are being developed, and if we can efficiently incorporate them into the human body, we can potentially improve the function and structure of proteins. The quadruplet codon system is proving to be the best option for this purpose, and further experiments and studies should be carried out to identify its full potential.
ACKNOWLEDGEMENTS
I would like to thank my mentor, Ryan Boyman, who works as a healthcare consultant at Simon-Kucher. He acted as my guide in narrowing down my research to a specific topic, helping collate sources and giving feedback on my work. I am very grateful to him for all the help and guidance I received. I would also like to acknowledge ACS Publications as well as John Wiley and Sons for granting permission to use these figures.
REFERENCES
- Wagner, I. and Musso, H. New naturally occurring amino acids. Angewandte Chemie International Edition in English, 22(11), pp.816-828 (1983).
- Noren, C. J., Anthony-Cahill, S. J., Griffith, M. C., and Schultz, P. G. A General Method For Site-Specific Incorporation Of Unnatural Amino Acids Into Proteins. Science 244, no. 4901, 182-188 (1989).
- Mayer, C. Selection, Addiction And Catalysis: Emerging Trends For The Incorporation Of Noncanonical Amino Acids Into Peptides And Proteins In Vivo. Chembiochem 20, no. 11, 1357-1364 (2019).
- Lopatniuk, M., Myronovskyi, M., and Luzhetskyy, A. Streptomyces albus: A New Cell Factory for Non-Canonical Amino Acids Incorporation into Ribosomally Synthesized Natural Products. ACS Chemical Biology, 12(9), 2362-2370 (2017).
- Ma, J. et al. Versatile strategy for controlling the specificity and activity of engineered T cells. Proceedings Of The National Academy Of Sciences, 113(4), E450-E458 (2016).
- Elsässer, S., Ernst, R., Walker, O., and Chin, J. Genetic code expansion in stable cell lines enables encoded chromatin modification. Nature Methods, 13(2), 158-164 (2016).
- Xue, L., Prifti, E., and Johnsson, K. A General Strategy for the Semisynthesis of Ratiometric Fluorescent Sensor Proteins with Increased Dynamic Range. Journal Of The American Chemical Society, 138(16), 5258-5261 (2016).
- Chin, J. Expanding and reprogramming the genetic code. Nature, 550(7674), 53-60 (2017).
- Yourno, J., and Kohno, T. Externally Suppressible Proline Quadruplet CCCUU. Science, 175(4022), 650-652 (1972).
- Bossi, L., and Roth, J. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell, 25(2), 489-496 (1981).
- Yourno, J. Externally Suppressive +1 “Glycine” Frameshift: Possible Quadruplet Isomers for Glycine and Proline. Nature New Biology, 239(94), 219-221 (1972).
- Kohno, T., and Roth, J. A Salmonella frameshift suppressor that acts at runs of a residues in the messenger RNA. Journal Of Molecular Biology, 126(1), 37-52 (1978).
- Moore, B., Persson, B., Nelson, C., Gesteland, R., and Atkins, J. Quadruplet codons: implications for code expansion and the specification of translation step size. Journal Of Molecular Biology, 302(1), 281 (2000).
- Anderson, J. C. et al. An expanded genetic code with a functional quadruplet codon. Proceedings Of The National Academy Of Sciences, 101(20), 7566-7571 (2004).
- Hankore, E. et al. Genetic Incorporation of Noncanonical Amino Acids Using Two Mutually Orthogonal Quadruplet Codons. ACS Synthetic Biology, 8(5), 1168-1174 (2019).
- Niu, W. An Expanded Genetic Code in Mammalian Cells with a Functional Quadruplet Codon. ACS Chemical Biology, 8(7), 1640-1645 (2013).
- Gubbens, J., Kim, S., Yang, Z., Johnson, A., and Skach, W. In vitro incorporation of nonnatural amino acids into protein using tRNACys-derived opal, ochre, and amber suppressor tRNAs. RNA, 16(8), 1660-1672 (2010).
- Mukai, T. et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Research, 38(22), 8188-8195 (2010).
- O’Donoghue, P. et al. Near-cognate suppression of amber, opal and quadruplet codons competes with aminoacyl-tRNAPyl for genetic code expansion. FEBS Letters, 586(21), 3931-3937 (2012).
- Heinemann, I. et al. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Letters, 586(20), 3716-3722 (2012).
- Zeng, Y., Wang, W., and Liu, W. Towards Reassigning the Rare AGG Codon in Escherichia coli. Chembiochem, 15(12), 1750-1754 (2014).
- Fischer, E. et al. New codons for efficient production of unnatural proteins in a semisynthetic organism. Nature Chemical Biology, 16(5), 570-576 (2020).
- Chatterjee, A., Lajoie, M., Xiao, H., Church, G., and Schultz, P. A Bacterial Strain with a Unique Quadruplet Codon Specifying Non-native Amino Acids. Chembiochem, 15(12), 1782-1786 (2014).
- Rackham, O., and Chin, J. A network of orthogonal ribosome·mRNA pairs. Nature Chemical Biology, 1(3), 159-166 (2005).
- Chen, I., and Schindlinger, M. Quadruplet codons: One small step for a ribosome, one giant leap for proteins. Bioessays, 32(8), 650-654 (2010).
- Wang, K., Neumann, H., Peak-Chew, S., and Chin, J. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nature Biotechnology, 25(7), 770-777 (2007).
- Neumann, H., Wang, K., Davis, L., Garcia-Alai, M., and Chin, J. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature, 464(7287), 441-444 (2010).
Written by Nandika Mishra
Seventh College, Molecular and Cellular Biology, Class of 2025