INTRODUCTION
Despite many prevention efforts, foodborne illnesses remain a serious global health threat.¹ The Centers for Disease Control and Prevention (CDC) recently reported that approximately 48 million people in the United States become ill with foodborne infections annually; this number includes both hospitalizations and deaths.² While there are numerous pathogens known to cause foodborne illnesses, Salmonella continues to be one of the leading causes, infecting a wide range of hosts through contaminated food and water. Healthy individuals recover within a few days, but those with weak or compromised immune systems–children, the elderly, and individuals with chronic disease–experience severe infection, or salmonellosis.² In order to improve treatment and prevention of salmonellosis, it is crucial to examine the host immune response to Salmonella infection.
SALMONELLA SECRETES BACTERIAL EFFECTOR PROTEINS FOR ENTRY AND SURVIVAL IN HOST CELL
During the infection process (Figure 1), Salmonella will depend on its Type III secretion system (T3SS) to produce bacterial effector proteins.³ These proteins are encoded by genes that fall within large gene cassettes known as Salmonella pathogenicity islands (SPIs).³ The effector proteins allow the bacteria to permeate the host tissue, subdue the host immune system, and to survive and replicate inside the host cell.3,4 Salmonella first invades epithelial cells by expressing effector protein genes such as SopB, SopE, and SopE2 within the Salmonella pathogenicity island 1 (SPI-1). Once Salmonella has crossed the epithelial lining, the pathogen is engulfed by phagocytes such as macrophages in the lamina propria, which are thin layers of connective tissue beneath the epithelium that form part of the mucous membrane. As it enters the mucous membrane, Salmonella encodes for another set of effector proteins in the Salmonella pathogenicity island 2 (SPI-2).
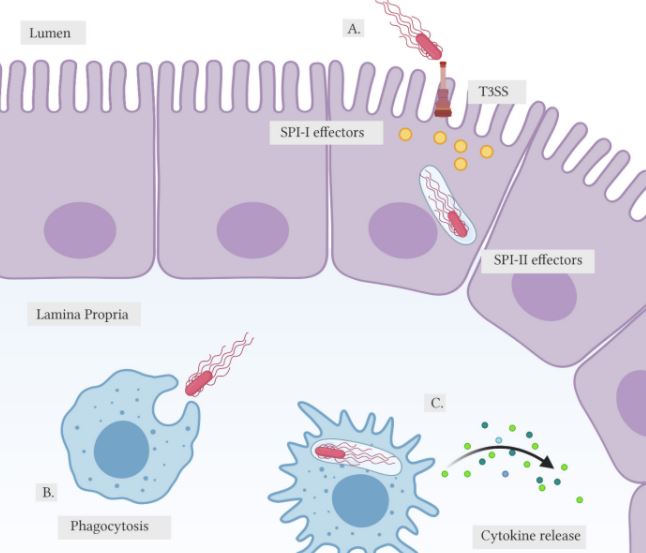
(A) Salmonella employs T3SS to inject SPI-I effector proteins, promoting invasion into the host epithelial cell. Upon entry, Salmonella will express SPI-II effector proteins to increase bacterial survival by forming the SCV.
(B) In the lamina propria, Salmonella will encounter macrophages where they will be engulfed via phagocytosis.
(C) Once engulfed, the macrophage will secrete cytokines to activate the host inflammatory response. Internalized Salmonella will again release SPI-II effectors to evade immune response. (created with biorender.com)
RECOGNITION OF BACTERIAL PATHOGENS AND ACTIVATION OF HOST IMMUNE RESPONSE
When a macrophage encounters a pathogen such as Salmonella, the macrophage detects the pathogen through specialized cell surface receptors known as pattern recognition receptors (PRRs). These receptors sense foreign ligands such as pathogen-associated molecular patterns (PAMPs), which contain
molecular motifs conserved within a group of microbes. Upon pathogen recognition, PRRs induce the activation of downstream signaling cascades in the macrophage, which then mounts an appropriate immune response.³ In a recent study, Das et al. identified Brain Angiogenesis Receptor 1 (BAI1) as a PRR that binds to lipopolysaccharides (LPS) of gram-negative bacteria.³ Upon binding, BAI1 recruits engulfment and cell motility protein 1 (ELMO1), a cytosolic microbial sensor protein that ultimately leads to the reorganization of the actin cytoskeleton to allow for the engulfment of certain pathogens.³
SOME PATHOGENIC BACTERIA EVADE IMMUNE RESPONSE THROUGH HIJACKING HOST CELL MACHINERY
Once the bacteria are internalized by a macrophage, the fate of the bacterium depends on its virulence factor or its ability to infect the host. Non-pathogenic bacteria are internalized into a compartment known as the early phagosome, which is formed through the invagination of the plasma membrane. Soon this compartment undergoes swift maturation into a late phagosome through a succession of membrane trafficking events. The phagosome then fuses with the lysosome that contain different hydrolytic enzymes to form the phagolysosome, where the internalized bacteria are eventually degraded. However, pathogenic bacteria such as Salmonella can avoid this fate by disrupting the trafficking mechanisms within the macrophage in order to survive and replicate. To protect itself, the internalized pathogen redirects the maturation of the phagosome to create a modified phagosome known as the Salmonella containing vacuole (SCV)., The Salmonella bacteria replicate inside the SCV, which has many features of the late endosome, including maintaining an acidic pH essential for the induction of SPI-2 T3SS bacterial effector proteins. In addition to this, Salmonella will block the fusion of the lysosome with the SCV and interactions with the endocytic pathway to prevent degradation of the bacterium.
BACTERIAL EFFECTOR PROTEINS INTERACT WITH CYTOSOLIC MICROBIAL SENSOR ELMO1 THROUGH A CONSERVED WXXXE MOTIF TO DISRUPT THE HOST INFLAMMATORY RESPONSE
Our recent findings indicate that ELMO1 assists macrophages in sensing pathogens and commensals to predict pathogenic infection through differential regulation of the host immune response. Considering that Salmonella bacterial effector SifA is important in maintaining SCV integrity for bacterial survival through disruption of the host immune response, our lab wanted to assess whether SifA interacts with ELMO1 to control pathogenesis.
Previous studies have found that ELMO1 interacts with the Shigella effector protein IpgB1, which is involved in entry into host cells through membrane ruffling. Since then, several known T3SS effectors including Salmonella SifA and SifB have been categorized into a single family of effector proteins that all contain a common WxxxE motif. Following the original classification, our lab has used BLAST to identify more enteric bacterial proteins that share this common motif; we found that these effector proteins are present in pathogenic bacteria but not in commensals (Figure 2). Moreover, preliminary studies have found that mutations in the WxxxE motif in Salmonella SifA affect its interaction with ELMO1.³,â·

A BLAST search of amino acid sequences in enteric pathogenic bacteria exhibit homologues that share a common amino acid sequence Trp-xxx-Glu or WxxxE motif (image courtesy of Das Lab).
The interaction between ELMO1 and other WxxxE effector proteins from Shigella and E.coli suggests that ELMO1 plays an important role in pathogenesis. Further research is needed to evaluate the interaction of ELMO1 with bacterial effectors secreted by pathogenic bacteria and to elucidate how these pathogens can hijack the host signaling machinery and impact the host inflammatory response.
REFERENCES
- Nyachuba D. G. (2010). Foodborne illness: is it on the rise?. Nutrition reviews, 68(5), 257—269. https://doi.org/10.1111/j.1753-4887.2010.00286.
- Centers for Disease Control and Prevention. (2020). Foodborne Germs and Illnesses. Retrieved from https://www.cdc.gov/foodsafety/foodborne-germs.html
- Das, S., Sarkar, A., Choudhury, S. S., Owen, K. A., Castillo, V., Fox, S., Eckmann, L., Elliott, M. R., Casanova, J. E., & Ernst, P. B. (2015). ELMO1 has an essential role in the internalization of Salmonella Typhimurium into enteric macrophages that impacts disease outcome. Cellular and molecular gastroenterology and hepatology, 1(3), 311—324. https://doi.org/10.1016/j.jcmgh.2015.02.003
- Ehrbar, K., Friebel, A., Miller, S. I., & Hardt, W. D. (2003). Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. Journal of bacteriology, 185(23), 6950—6967. https://doi.org/10.1128/jb.185.23.6950-6967.2003
- Alto, N. M., Shao, F., Lazar, C. S., Brost, R. L., Chua, G., Mattoo, S., McMahon, S. A., Ghosh, P., Hughes, T. R., Boone, C., & Dixon, J. E. (2006). Identification of a bacterial type III effector family with G protein mimicry functions. Cell, 124(1), 133—145. https://doi.org/10.1016/j.cell.2005.10.031
- Bulgin, R., Raymond, B., Garnett, J. A., Frankel, G., Crepin, V. F., Berger, C. N., & Arbeloa, A. (2010). Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infection and immunity, 78(4), 1417—1425. https://doi.org/10.1128/IAI.01250-09
- Sayed, I. M., Suarez, K., Lim, E., Singh, S., Pereira, M., Ibeawuchi, S. R., Katkar, G., Dunkel, Y., Mittal, Y., Chattopadhyay, R., Guma, M., Boland, B. S., Dulai, P. S., Sandborn, W. J., Ghosh, P., & Das, S. (2020). Host engulfment pathway controls inflammation in inflammatory bowel disease. The FEBS journal, 287(18), 3967—3988. https://doi.org/10.1111/febs.15236
- Handa, Y., Suzuki, M., Ohya, K., Iwai, H., Ishijima, N., Koleske, A. J., Fukui, Y., & Sasakawa, C. (2007). Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nature cell biology, 9(1), 121—128. https://doi.org/10.1038/ncb1526
- Mellouk, N., & Enninga, J. (2016). Cytosolic Access of Intracellular Bacterial Pathogens: The Shigella Paradigm. Frontiers in cellular and infection microbiology, 6, 35. https://doi.org/10.3389/fcimb.2016.00035
WRITTEN BY SAREH KARIMILANGI
M.S. Biology, UC San Diego 2021, Das Lab, George Palade Laboratories for Cellular and Molecular Medicine
FROM SALTMAN QUARTERLY VOL. 18
To read the original version, please click here. To read the full version on our website, please click here. To read more individual articles, please click here.
