Successive generations of organisms may experience many changes in their physiology, behavior, and
development. While most traits, such as beak shape or coat color, can be perceived by the human
eye, the actual changes that produce variation in these traits occur at a molecular level.¹ These
molecular differences can be used in genetic engineering to confer adaptive advantages for organism
survival.² Researchers who study population genetics compare the genetics between organisms of
interest to understand the molecular forces driving evolutionary change. These studies generally
involve assessing the differences in DNA in a species over time and correlating these differences with
variations in phenotype and variable survival rates.³
The Rennison Lab at the Ecology, Behavior and Evolution Section within the Division of Biological Sciences at UC San Diego aims to examine model systems that highlight the genetic differences in evolution. Dr. Andreas Haerer, a post-doctoral researcher in the Rennison Lab, focuses on studying alterations in the gut microbiome and how they affect ecology and evolution of the host. He has published a paper characterizing the relationship between gut microbiome changes and the parallel evolution of the hosts. Parallel evolution is the process by which similar traits arise independently of one another.
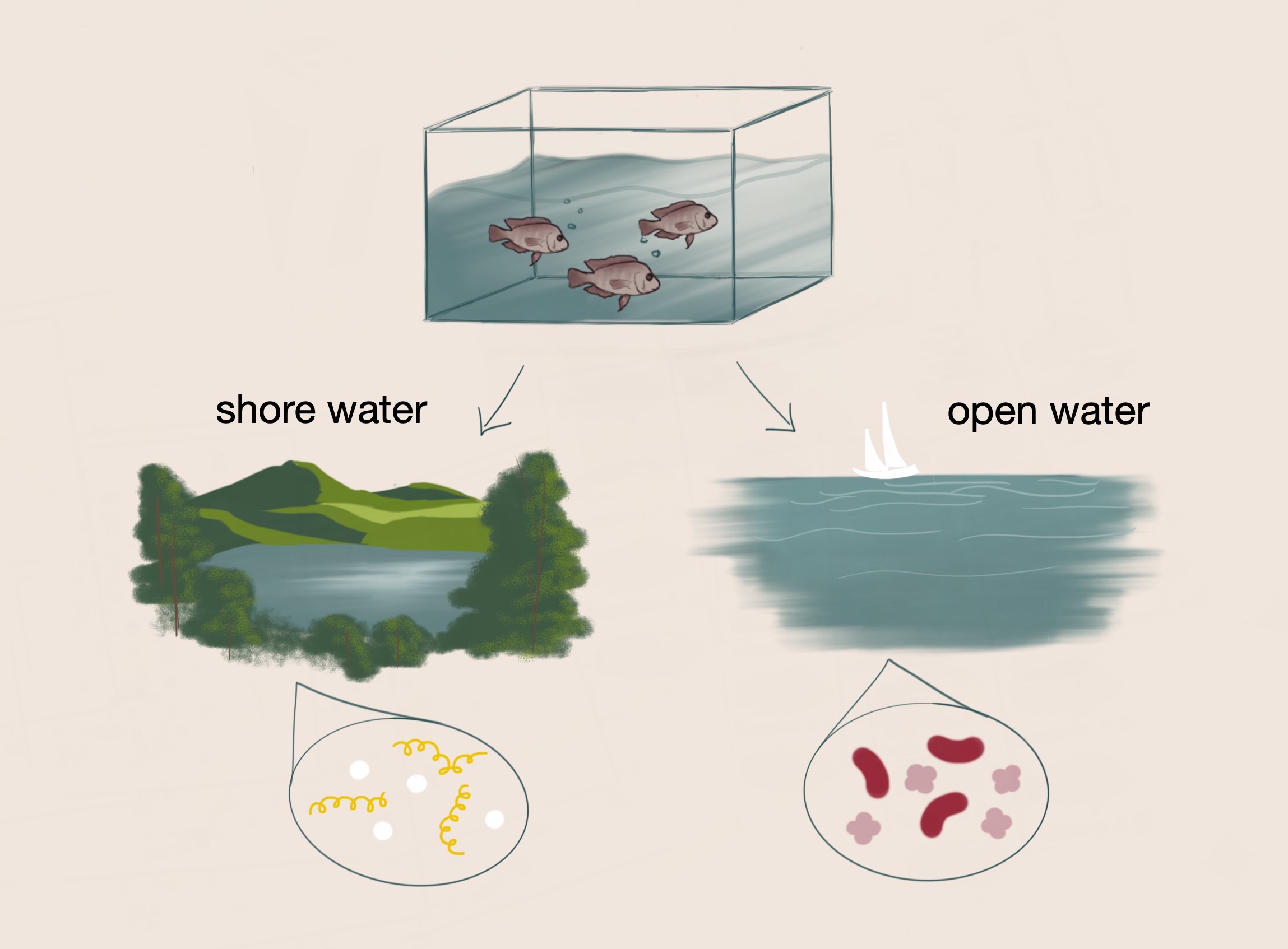
Fish that initially originated from the same environment diverged into separate lake environments with distinct conditions. Dr. Haerer hypothesized that parallel changes in diet and habitat are associated with parallel changes in the gut microbiomes of the fish.
One of his studies examines the relationship between diets of different fish species and the differences in their gut microbiomes. In his controlled experiment, Dr. Haerer aimed to identify changes in diet and habitat that would result in changes of the gut microbiome of cichlid fish. Initially, the ancestors of the cichlid fish colonized the same lake. However, the fish were eventually separated into two different lake environments with variable conditions. Within these lakes, ecological divergence and speciation occurred as a result of adaptation to different niches. Over several generations, the shore water fish and the open water fish developed distinct gut microbiomes. The fish in these respective environments additionally have habitat-specific diets which are also associated with distinctive gut microbiomes. Diet-specific microbes have the potential to modulate digestion, intake of food, and health; as such, these respective microbes may confer selective advantages on these fish, such as more efficient processing of food and resistance to disease.³ From these initial observations, Dr. Haerer hypothesized that parallel changes in the gut microbiomes of the fish across the lakes are associated with parallel changes in diet and habitat.
To test this hypothesis, Dr. Haerer and his team traveled to Nicaragua to collect fish from local lakes as samples for the study. Dr. Haerer and colleagues used a pair-wise study to analyze the fish in pairs: fish from each lake were compared to control fish in external analyses to assess whether the lake and diet resulted in a significant difference in microbial composition between the fish. The team used 16s rRNA sequencing to identify microbes based on the unique sequences of the 16s rRNA gene, which is highly conserved across microbial species. In addition to collecting fish samples from Nicaragua, Dr. Haerer obtained water samples to ensure the respective microbes in the surrounding environment of the fish could be determined. After sequencing the fish gut samples to characterize the microbial composition of the fish, Dr. Haerer analyzed the fish diets through stable isotope ratio analysis. This type of analysis identifies nitrogen and carbon isotope data, which can indicate where the fish feed in the lake in a three month period–by the shore versus in the open water.
From the analyses, Dr. Haerer was able to identify a positive correlation between differences in diet and the variation in gut microbial composition. While Dr. Haerer found differences in the gut microbiota across species of fish and correlations between the gut microbiome and diet, his team did not find strong evidence for parallelism of gut microbiota changes. As such, he concluded that perhaps there was insufficient differentiation in diet to promote alterations in gut microbial composition.
The research conducted by the Rennison Lab contributes to the understanding of the evolution of organismal traits. In the future, the Rennison Lab hopes to continue pioneering research to explore the mechanisms by which microbes can provide evolutionary benefits to their hosts and how microbes subsequently impact their hosts’ evolutionary trajectory.
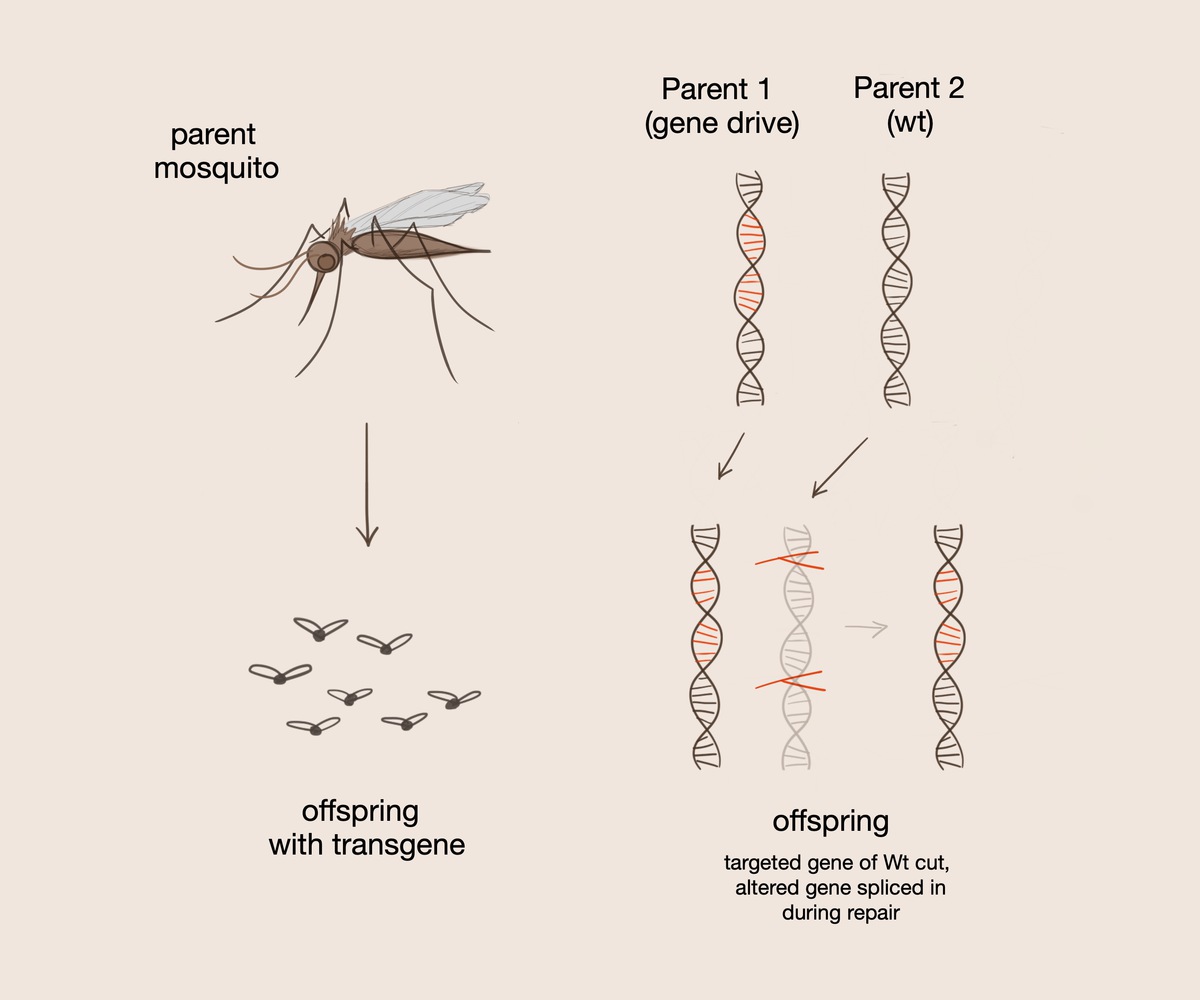
A gene drive serves to convert heterozygotes to homozygotes for the gene of choice such that the desired gene is always passed on to the offspring. The gene drive is introduced into selectively mated organisms. It contains guide RNA, which tags certain sections of DNA to be cut. The Cas9 protein subsequently cuts the double stranded DNA at a specific recognition sequence.
Understanding the genetic changes responsible for phenotypic change can be important for preserving a species or aiding in preventing the spread of disease. The Akbari Lab at UC San Diego is particularly interested in mitigating the transmission of malaria, which is often spread by mosquitoes. Dr. Andie Smidler, a researcher in the Akbari Lab, is currently using population genetics to combat malaria by making mosquitoes incapable of transmitting the disease. The Akbari Lab uses the CRISPR/Cas9 system to study malaria. CRISPR/Cas9 is a form of genetic engineering technology that enables the genome to be edited or altered.
Although utilizing the novel CRISPR/Cas9 system opens a myriad of possibilities, there are many challenges to genetic engineering research, including ethical considerations, practical experimental issues, and environmental effects. The effects of releasing organisms genetically modified with CRISPR/Cas9 into the environment are relatively unstudied. As such, the researchers took ethical and practical considerations into account when designing their experiment. In their studies, researchers at the Akbari Lab found that there are many different ways to reduce the spread of malaria.
One of these methods involves introducing mosquitoes to a gene that has the ability to kill the malaria parasite; however, in this case, the researchers found that all mosquitoes must be introduced to this specific gene in order to eradicate malaria. In order to increase gene transmission, the researchers introduced a gene drive, a system which uses aspects of the CRISPR/Cas9 system on a larger scale, into the offspring of selectively mated mosquitos. The gene drive itself contains the Cas9 protein, which cuts the double stranded DNA at a specific recognition sequence. This system also contains the guide RNA, which tags certain sections of DNA to be cut, and homology arms, which facilitate chromosome recombination and interactions between the gene drive and organism DNA. A gene drive serves to convert heterozygotes to homozygotes for the gene of choice such that the desired gene is always passed on when present.
Another technique for eradicating malaria involves a sterilization technique to suppress mosquito populations. Under normal circumstances, female mosquitoes only mate once and are unable to reproduce after that initial mating. By sterilizing the male mosquitoes through targeted radiation, thereby ensuring no offspring are produced, Dr. Smidler hypothesized that future generations of mosquitoes would be suppressed, which would reduce the spread of malaria. Dr. Smidler also found that the CRISPR/Cas9 genetics engineering system can be used to cut the gene associated with fertility in male mosquitoes, thus sterilizing them.
However, this system also brings up ethical concerns, as eliminating the progression of a species may disrupt natural selection and may have unintended consequences on the ecosystem. These consequences may include species extinction, the elimination of a source of food for other organisms, and the proliferation of pests that mosquitoes may consume. Moreover, there remain concerns regarding the safety and feasibility of genomically edited organisms. Organisms with altered genes may not be able to survive in their natural habitat and may cause harmful effects on the environment and other organisms.â·
Additionally, simply sterilizing male mosquitoes may not address the issue of malaria completely, as female mosquitoes still retain the ability to bite others and spread malaria on their own. To combat this problem, the Akbari Lab plans to use a gene drive to render the gene associated with malaria nonfunctional in female mosquitoes, in order to reduce the spread of this deadly disease. By mitigating malaria with a multifaceted approach, the Akbari Lab hopes to eliminate malaria by reducing the genetic material that is passed onto offspring and preventing transmission of malaria from female mosquitoes.
Genetic variation can be used in a broad scope of applications, including eradication of progressive diseases. By combining studies of genetic, immunological, and microbial differences with methods that harness these findings on a larger scale, we can use population genetics to reduce both deadly diseases and their associated mortalities. We can also increase the survival rates of certain protected species by understanding their evolutionary trajectories. By examining the changes and hereditary nature of the microbiomes in fish species over time, Dr. Haerer and the Rennison Lab were able to identify the molecular mechanisms that affected ecosystem dynamics. Similarly, the Akbari Lab was able to harness molecular and population genetics mechanisms to investigate methods through which ecosystem dynamics can be altered.
REFERENCES
- Bonsall M, Charlesworth B. 2010. Genetics and the causes of evolution: 150 years of progress since Darwin. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 365(1552), 2427—2429.
- Karalis DT, Karalis T, Karalis S, Kleisiari AS. 2020. Genetically Modified Products, Perspectives and Challenges. Cureus, 12(3), e7306.
- Valdes AM, Walter J, Segal E, Spector TD. 2018. Role of the gut microbiota in nutrition and health. BMJ, 361:k2179.
- Zimmo S, Blanco J, Nebel S. 2012. The Use of Stable Isotopes in the Study of Animal Migration. Nature Education Knowledge 3(12):3.
- Degner EC, Harrington LC. 2016. Polyandry Depends on Postmating Time Interval in the Dengue Vector Aedes aegypti. The American Journal of Tropical Medicine and Hygiene, 94(4), 780—785.
- Conrad DF, Hurles ME. 2007. The population genetics of structural variation. Nature Genetics, 39 (7 Suppl), S30—S36.
- Caplan AL, Parent B, Shen M, Plunkett C. 2015. No time to waste–the ethical challenges created by CRISPR. EMBO Rep, 16:1421-1426.
WRITTEN BY SHRUTI MAGESH
Shruti is a Human Biology Major from Roger Revelle College. She will be graduating in 2024.
FROM SALTMAN QUARTERLY VOL. 18
To read the original version, please click here. To read the full version on our website, please click here. To read more individual articles, please click here.
