In 2006, Kazutoshi Takahashi and Shinya Yamanaka published a study that won them a Nobel Prize for their breakthroughs in generating induced pluripotent stem cells (iPSCs) from already-existing matured cells in mice. In 2007, they took their Nobel-winning research a step further, generating pluripotent stem cells from human dermal fibroblasts. These fibroblasts are a common type of somatic, or non-reproductive, cell found in the skin. From fibroblasts, Yamanaka and Takahashi successfully generated iPSCs from them. Turning skin cells into stem cells created an accessible method for the regeneration of desired stem cells. Their research revolutionized many fields in the biological sciences, including neuroscience. It created the foundation for future studies that would use these very methods to study a variety of different neurological disorders. Dr. Hiruy Meharena, an expert in this field joins us in an interview to discuss the progression and possibilities of stem cell research in neuroscience and beyond below.
Stem cells are cells that have not yet differentiated, or changed, into cells with unique structures and unique functions. Differentiation can be triggered by several factors, such as environmental cues. Stem cells within the central nervous system, known as neural stem cells (NSCs) turn into different kinds of neural cells. At the pluripotent stem cell stage, or the stage where the stem cell is still immature, differentiation into multiple types of matured cells can be highly controlled. Having this controlled differentiation into specific neural cells such as neurons and glial cells– a type of neuron-support cell– allow many scientists to use stem cells to grow cells as needed. This technique has been included in studies to advance knowledge on neurological disorders such as schizophrenia, Parkinsons, and Alzheimers, among other disorders.
Schizophrenia:
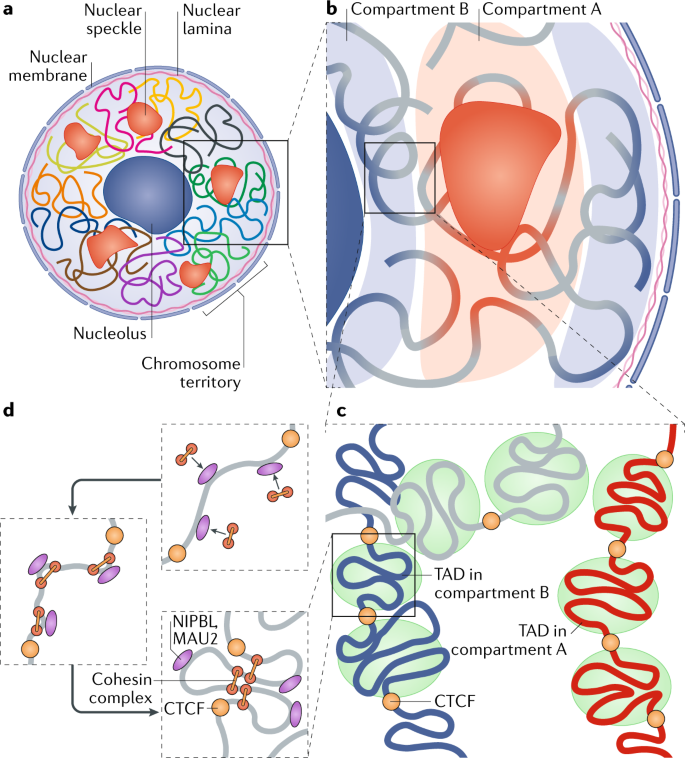
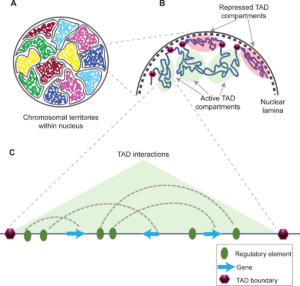
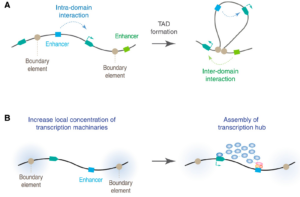
Schizophrenia is both a psychiatric and neurological disorder that is largely influenced by an individual’s genes. A 2019 study used stem cell technology to predict possible genetic contributions to schizophrenia pathology. This disorder is a particularly complex one to study because contributing mutations lie in the noncoding regions of the gene, or the parts of the gene that are not directly expressed but can influence the expression of expressed coding DNA regions. These noncoding regions include promoters, enhancers, and repressors, which control the rate or amount of gene expression through interactions with different coding and noncoding regions in the genome. When an enhancer interacts with a promoter region, gene expression is amplified, but a repressor decreases the rate of gene expression. Furthermore, chromatin—genetic material containing DNA and proteins within each cell—takes on a 3-dimensional and nonlinear orientation known as the 3D genome (3DG). The nonlinear 3DG includes chromatin loops known as TAD loops which help interactions within genes. TAD loops allow promoters, enhancers, and repressors otherwise kilobases apart on a linear line to become within proximity in a loop. This then allows these noncoding regions to interact with each other. However, this long-distance gene interaction is exactly what makes the 3DG and schizophrenia so difficult and complex to study: the presence of TAD loops in the 3DG means that non-coding regions are able not only to interact with the regions closest to them, but also the regions that would otherwise be much, much farther away.
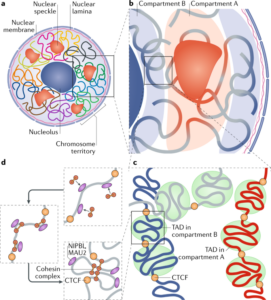
Now, having mutations in the noncoding regions that influence gene expression in the 3DG could mean that the mutations influence more than a handful of coding regions. If the DNA took a linear conformation, only the promoters and enhancers relatively close to each other would interact and cause gene expression. This would make mutations in noncoding regions of the DNA, such as promoter regions, potentially far simpler to identify, and these mutations would not reach as far across the gene. To study genetic interaction in the 3DG, the researchers used a technology called Hi-C to track connections of different genomic regions across the entire genome. Tracking interaction across the whole genome eliminates the bias of picking and choosing which regions to study. This study used Hi-C to focus on presence and intensity of interactions between different regions of the gene with noncoding regions that are known to be at risk for schizophrenia.
From two non-schizophrenic participants, the authors generated iPSCs which differentiated into neural progenitor cells (NPCs), glutaminergic neurons, and astrocytes. NPCs are a type of stem cell that, unlike normal stem cells, have a more targeted range of mature cells they can differentiate into. The glutaminergic neurons of this study are excitatory neurons that keep “passing along the message” to other neurons instead of stopping the chain. Astrocytes are a type of glial– or non-neuronal–cell that specifically resides in the central nervous system (CNS), which includes the brain. Astrocytes are characterized by their star-like formation, regulating the extracellular environment of CNS neurons. By using Hi-C to observe genetic interactions within each specific type of cell, the researchers attempted to derive exactly how schizophrenia might be influenced by interactions between noncoding regions.
Upon using Hi-C to observe the interactions between noncoding regions and TAD amounts, the authors found that of the three cell types, neurons had significantly fewer TAD loops compared to astrocytes and NPCs. However, TAD sizes were larger in neurons. This implied that though there was a lower amount of loops, noncoding regions further away from each other could interact in neurons that couldn’t in astrocytes or NPCs. To further visualize the interactions between noncoding regions of the gene, specifically of at-risk schizophrenia genes, the authors created a heat map to display what Hi-C observed. Heatmaps use color, such as red, to indicate the interaction between gene regions. If there were no interaction, there would be no color shown on the heatmap. In the heat map they created, the more intense the color, the more interaction within or contacting the specific at-risk schizophrenia locus in this focus. Neurons had the greatest amount and intensity of protocadherin (PCDH) interactions and risk locus connection. PCDH are specific molecules that influence neuronal development. Through these and other analyses, it was determined that neurons have the highest risk-contact within their genes compared to astrocytes and NPCs, with schizophrenia-causing at-risk promoters possibly interacting with more genetic regions. From this study, the authors determined that interactions among these risk regions may theoretically contribute to schizophrenia development. This is just one result of the many experiments these researchers conducted in their attempt to understand how interactions within the genome might contribute to schizophrenia.
Schizophrenia is one of many neurological disorders whose research benefited from the stem cell technology invented by Takahashi and Yamanaka. Though this research takes a more theoretical approach to the complex disorder, it provides valuable insight and a promising plan to move forward in our understanding of this neuropsychiatric disorder. Many other disorders are being studied using stem cell technologies, including multiple sclerosis, Parkinson’s, Huntington’s, Alzheimers, and autism.
Below is an interview with Dr. Hiruy Meharena, principal investigator of the Meharena Lab at UC San Diego and professor of UCSD’s undergraduate BIPN 194 and graduate BGGN 284 in the seminar on Stem Cell Models in Neuroscience. He researches the cell-type specific gene-networks and molecular indications behind the development of neurodevelopmental disorders such as Autism Spectrum Disorder and Down syndrome using iPSCs.
SQ: How might stem cells teach us about neurological and neurodevelopmental disorders that also rely on environmental and social factors or other factors than genetic code?
Meharena: I think one of the interesting things about stem cells is because they’re human-derived. You’d be able to provide them with different environmental factors. These environmental factors have to be chemical-based. For example, if you wanted to study how alcohol impacts neurodevelopment, you can start generating a brain in a dish, and then add some level of alcohol and see how that impacts the development of the brain. You can also study environmental factors such as infectious disease or maternal stress. Stress is basically a chemical compound at the end of it. It’s like adding hydrogen peroxide to look at mitochondrial stress and look at how the brain develops.
SQ: How might advancements in stem cell research transform the understanding of neurological disorders and in the clinical application?
Meharena: One of the biggest challenges that we’ve had in the past was that all neurological studies were done on mice or other rodents. Humans and rodents are not similar or the same, so there were a lot of gaps that we were missing in our knowledge about human neurological systems. This includes at the molecular and cellular level, just understanding through development, neuronal function and glia… how are they not only functioning at baseline but how are they interacting with other cell types in their system. Human stem cells now allow us to do that. One of the most difficult things in my opinion has been [ability] to study neurodevelopment. You cannot track neurodevelopment in a pregnant mom the way you’d be doing it with mice, in that we sacrifice mice at different time points and can extract the brain to try to understand what are the specific changes that are happening with high spatial temporal resolution. But now that we have organoids, we can at least somewhat recapitulate that.
(Organoids are 3D tissue samples produced from stem cells)
SQ: What do you think is the most beneficial aspect of combining improvements in stem cell technology with clinical practice?
Meharena: I think we’d need to start with some drawbacks or limitations of stem cell technology, especially in the context of the brain. All the models that we have would allow us to either generate the specific cell types and understand them in a 2D model which is unrealistic– we don’t exist in a 2D model. We’ve advanced to trying to make 3D models. However, the 3D models originate from neural progenitor cells; our brains are constructed of way more than that. We have… cells that have not been fully incorporated into the 3D models that we have today. We have started trying to integrate that, but we’re not there yet. There’s a significant limitation in that regard. The biggest problem is when an organism develops, there are all these different organs [other than the brain]; there are all of the factors that the other organs release, which is not available in that [stem cell] system]. There is a lot of drawback [to this technology] in that we’re still not able to mimic human brain or brain development in our stem cell systems.
There are no FDA-approved stem cell therapies today, but it’s something that’s in progress and the most advanced in that area is neurodegenerative disorders.
(Understanding the limitations of stem cell technologies is necessary to create new research methods or technologies that allow us to surpass them and produce even more effective and accurate results and applications.)
SQ: Do you foresee or are there any major advancements currently being made or considered for stem cell research that may improve its field?
Meharena: The major thing that I think will probably come in the near future is organ transplantation. One of the biggest challenges today in medicine is the limitation of the number of organs that are available for organ transplantation, and even after transplantation there’s rejection and so many different issues on compatibility. However, imagine a world where you can take a skin cell from the patient and make the organ that you need. There are several companies that are attempting to do this. This would be chimeric models with pigs where you take a human stem cell and try to generate the different organs that you need. I think this is probably where we will see some exciting new advancements.
(A chimeric model includes inserting pluripotent stem cells from one individual within a species to another, either of the same or different species, and allowing them to differentiate and grow.)
SQ: Can you tell us about any current research interests or projects that you’re doing right now?
Meharena: My lab is interested in intellectual disabilities… We utilize stem cells to generate organoids and look at how they develop… We have a model that we call “memory in a dish” which is matured neurons where we can activate them… with light… and look at the genetic and molecular changes that happen with neuronal activity. We’re trying to understand how autism and Down syndrome basically disrupt this process.
SQ: Do you have any advice for those interested in conducting research or learning about neurological disorders using stem cells, but may be daunted by the complexity of the topic?
Meharena: I think we’re in a really privileged position here at UCSD because we have the Sanford Center of Regenerative Medicine, and everyone in that building does stem cell research. There are a lot of different labs across campus that are utilizing stem cells, and it’s not just in the field of neuroscience and neurobiology but in cancer and other organ abnormalities, development, or function. There’re a lot of great opportunities. A great way to start is to volunteer in a lab. BIPN 194 [seminar class taught by Dr. Meharena] is a good place to start too.
SQ: Do you have any comments on this field of research that we haven’t covered so far?
Meharena: I will say that it’s a really exciting field. It has a lot of potential to grow, and with the ability to generate chimeric models we’re overcoming a lot of obstacles that have been limiting us in terms of making progress. I feel there’s going to be significant exciting avenues opening up soon. For that to happen, we need more people to be engaged and participating in research.
References:
https://pubmed.ncbi.nlm.nih.gov/16904174/
https://pubmed.ncbi.nlm.nih.gov/18035408/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5963504/
https://pubmed.ncbi.nlm.nih.gov/30545851/
https://www.frontiersin.org/articles/10.3389/fgene.2021.681259/full
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3149993/