By: Lauren Brumage | SQ Staff Writer 2016-2017
The medical advancement that has saved thousands of lives over the past century is now crumbling, leaving in its wake annual health care costs of $21 to $34 billion in the United States. Antibiotics, once a panacea for bacterial infections, are becoming increasingly ineffective as genes for resistance spread. The mortality rate for one antibiotic resistant infection, carbapenem-resistant enterobacteriacea (CRE), is an astonishing fifty percent. Unless novel antimicrobial therapies are implemented, mortality rates for antibiotic resistant infections will rise.
Antibiotic resistance stems from a variety of factors including some that are preventable. Worldwide phenomena such as overprescription and misuse of antibiotics facilitate natural selection for resistant strains of bacteria. In some countries, antibiotics are even available over the counter for citizens to self-medicate – a phenomenon that encourages improper dosage. Antibiotic use enhances the survivability of strains that have resistance genes over those that do not. Therefore, it is imperative to carefully select the proper antibiotic treatment plan for a given infection.
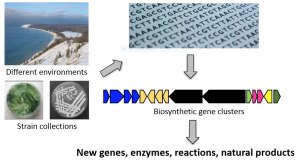
In spite of the resistance crisis, few pharmaceutical companies are actively conducting antibiotic discovery research due to tedious techniques and frequent failures. The revolutionary technology of genome mining may change this. Genome mining involves sequencing a bacteria’s genome and scanning it for “cryptic” biosynthetic clusters whose precise product is unknown (see Figure 1). Biosynthetic clusters contain genes for biosynthetic pathways that yield natural products, some of which have antibiotic properties. By analyzing these clusters, researchers at the University of Illinois, Urbana-Champaign found 19 new natural products in 4 years. Although further research needs to be conducted on the natural products discovered, the study establishes genome mining as an efficient method of discovering potential antibiotics.
However, doctors can bypass antibiotics entirely through exploiting bacteriophages, viruses that infect bacteria. Phage therapy has been used since the early 1900s in Eastern Europe. Each phage has a host range of bacteria species that it can infect, allowing doctors to select the appropriate phage for a given infection. Such specificity facilitates avoidance of dysbiosis, a condition frequently attributed to antibiotic use, wherein the patient’s helpful bacteria suffer. Furthermore, phage therapy is fairly cost-effective and has low risk for adverse side effects if the phage used has a lytic life cycle.
A study by Smith and Huggins of Houghton Poultry Research Station in Cambridgeshire on intramuscular and intracerebral Escherichia coli infections in mice showed that one dose of phage lysate combatted the infection better than four major antibiotics. Nevertheless, scientists must be careful as not all bacteriophages promulgate a positive outcome. Whereas lytic phages replicate and lyse their host shortly after infection, temperate phages integrate their genome with the host genome. Since this integration permits temperate phages to deliver toxin-encoding genes to the host bacteria, effectively augmenting the host’s arsenal, temperate phages are higher risk candidates for phage therapy.
Genome mining presents a promising frontier in a field that has seen few significant discoveries over recent decades. While it is undeniably beneficial to discover novel antibiotics, resistance to these compounds will arise. In the words of Dr. Arjun Srinivasan, an associate director at the Center for Disease Control, it is entirely plausible that “the end of antibiotics” is nigh. Consequently, it is imperative to pursue phage therapy and other novel methods alongside antibiotic discovery research. Broadening treatment horizons will save lives and reduce healthcare costs.
[hr gap=”0″]
Sources:
- Abedon, S. T., Kuhl, S. J., Blasdel, B. G., & Kutter, E. M. (2011, March/April). Phage treatment of human infections. Bacteriophage, 1(2), 66-85. doi:10.4161/bact.1.2.15845
- Antibiotic/Antimicrobial Resistance. (2015). Retrieved June 22, 2016, from https://www.cdc.gov/drugresistance/biggest_threats.html
- Childress, S. (2013, October 22). Dr. Arjun Srinivasan: We’ve Reached “The End of Antibiotics, Period”. Retrieved June 22, 2016, from http://www.pbs.org/wgbh/frontline/article/dr-arjun-srinivasan-weve-reached-the-end-of-antibiotics-period/
- Golkar, Z., Bagasra, O., & Pace, D. G. (2014). Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. The Journal of Infection in Developing Countries, 8(02). doi:10.3855/jidc.3573
- Michael, C. A., Dominey-Howes, D., & Labbate, M. (2014, September 16). The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Frontiers in Public Health, 2. doi:10.3389/fpubh.2014.00145
- Scheffler, R. J., Colmer, S., Tynan, H., Demain, A. L., & Gullo, V. P. (2012). Antimicrobials, drug discovery, and genome mining. Applied Microbiology and Biotechnology, 97(3), 969-978. doi:10.1007/s00253-012-4609-8
- Smith, H. W., & Huggins, M. B. (1982). Successful Treatment of Experimental Escherichia coli Infections in Mice Using Phage: Its General Superiority over Antibiotics. Microbiology, 128(2), 307-318. doi:10.1099/00221287-128-2-307
- Sybesma, W., & Pirnay, J. (2016). Silk route to the acceptance and re-implementation of bacteriophage therapy. Biotechnology Journal, 11(5), 595-600. doi:10.1002/biot.201600023
- The antibiotic alarm. (2013, March 14). Retrieved June 22, 2016, from http://www.nature.com/news/the-antibiotic-alarm-1.12579
- University of Illinois at Urbana-Champaign. (2015, September 8). Genome mining effort discovers 19 new natural products in four years. ScienceDaily. Retrieved June 22, 2016 from www.sciencedaily.com/releases/2015/09/150908133317.htm

